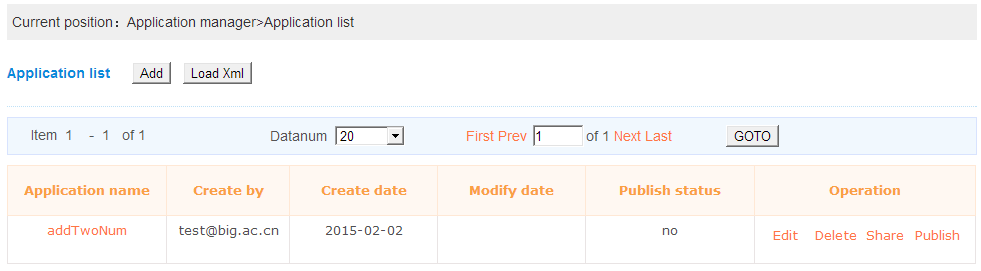
2.1 Application List
This page will show all the created applications and the shared applications from other persons of the current login user. User can edit,delete the application and share,publish applications to other persons.User click the “Add” button will open the web page of create one application and “Load Xml” will open the upload page of create one application.Note: If the edit operation has been done,user need to wait the application to be validated by BioCloud. If the application has been published,user can not delete it any more.
 Share an application:Users can share the application to other person,when the user login into the system the shared application can be used.The shared application only available for the shared users.
Share an application:Users can share the application to other person,when the user login into the system the shared application can be used.The shared application only available for the shared users.
 Publish an application:Users can publish the application immediately or publish at the given date.Click the “Browse” button, the application type dictionary will be loaded.When the application published, all the BioCloud users can use this application.
Publish an application:Users can publish the application immediately or publish at the given date.Click the “Browse” button, the application type dictionary will be loaded.When the application published, all the BioCloud users can use this application.

2.2 Create an application
Two ways are available to create an application in biocloud, one is the web form,the other is xml format.
1)By web form
When click “Add” under application list ,the page for create application will be show.All the items under this page are described here.
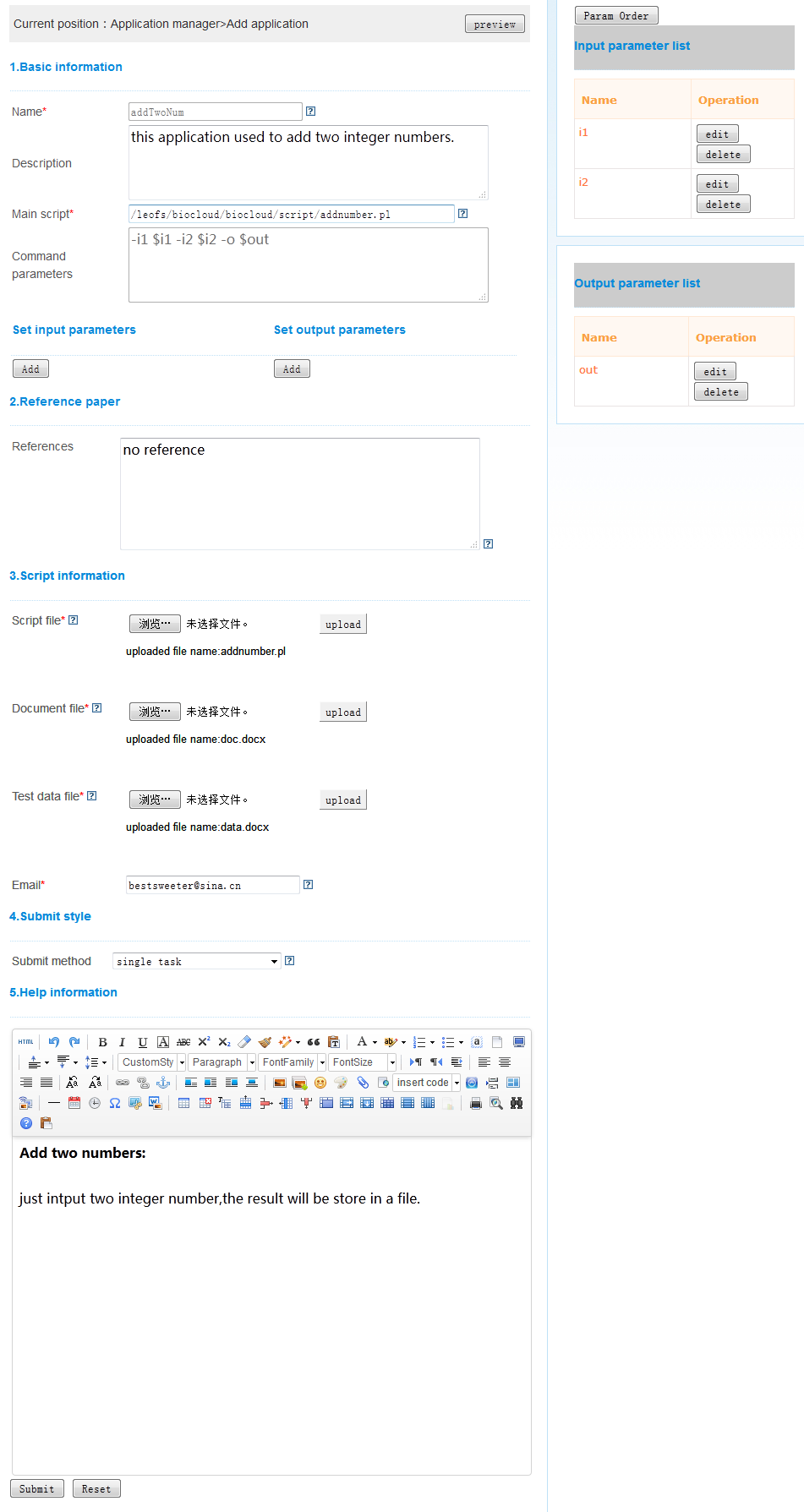
Description of the parameters as following table.
| Name | Description |
|---|---|
| Name | an unique application name,which consisted by letters(a-z,A-Z),numbers(0-9) and underline(_),such as add_number. Note:white space and other special letters are not allowed. |
| Description | a brief description about this application |
| Main script | a script used to active this application.The users need to provide the script running method except the detail path. Such as java -jar drawpic.jar perl drawpic.pl drawpic.sh |
| Command parameters | a read only field, used to show the input and output parameters which configured by users |
| Reference paper | several publish papers which used by this application |
| Script file | all the scripts used by this application.If the script file more than one file,users should packaged all of them into one file.If the script used some public softwares, users can describe them in the document file. |
| Document file | the document describe how to deploy this application under linux machine.If some system environment parameters need to configure,users must describe clearly here |
| Test data file: | the minimum test data used to test the application. |
| an email address used to contact the person if we deploy the application encounter some problems | |
| Submit method | two method provides, single task for applications which run on single machine;multiple tasks for applications which run under MPI environment. Biocloud use PBS to schedule task, users can set the node number,cpu number, running queue,running memory and running time. |
| Help | how to use this application.We suggest users provides information as detailed as possible. |
Set input parameters
For most command line, users can use command parameter and the default value to finish.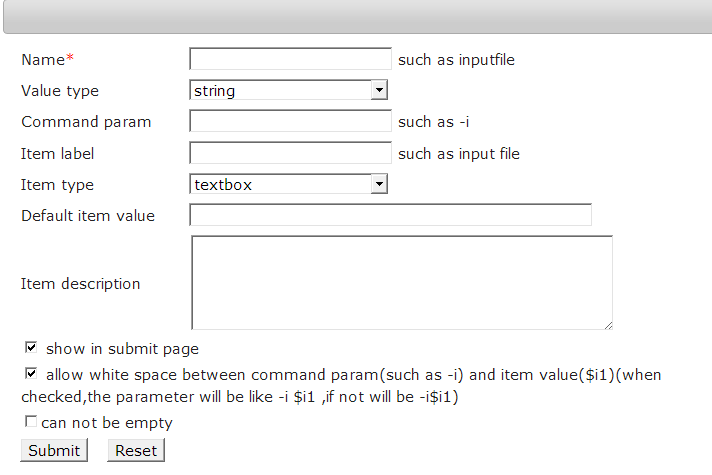
Description of the parameters as following table.
| Name | Description |
|---|---|
| Name | an unique parameter name in this application,which consisted by letters(a-z,A-Z),numbers(0-9) and underline(_),such as add_number. Note:white space and other special letters are not allowed. |
| Value type | the value type of this parameter,different value type will have different item type. |
| Command param | the command line prompt parameter, which mostly used in perl script,such as -i |
| Item label | a short label about this parameter. |
| Item type | HTML item used by this parameter |
| Default item value | a default value of this parameter,which will show by the chosen item type. |
| Item description | a brief description about this parameter |
| Show in submit page | if checked, it means this parameter will show in the submit page by the chosen item type. |
| Allow white space | if checked,it means allows white space between command parameter(-i) and the parameter(i1),such -i i1 |
Set output parameters
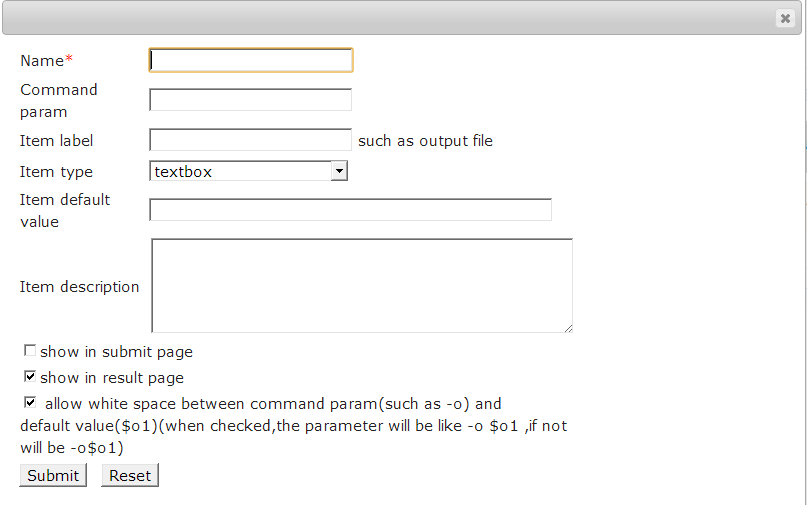
Description of the parameters as following table.
| Name | Description |
|---|---|
| Name | an unique parameter name in this application,which consisted by letters(a-z,A-Z),numbers(0-9) and underline(_),such as add_number. Note:white space and other special letters are not allowed. |
| Command param | the command line prompt parameter, which mostly used in perl script,such as -o |
| Item label | a short label about this parameter. |
| Item type | HTML item used by this parameter,only two types support textbox and checkbox |
| Default item value | a default value of this parameter,which will show by the chosen item type. |
| Item description | a brief description about this parameter |
| Show in submit page | if checked, it means this parameter will show in the submit page by the chosen item type. |
| Show in result page | if checked,it means this parameter will show in the result page when the task finish |
| Allow white space | if checked,it means allows white space between command parameter(-o) and the parameter(o1),such -o o1 |
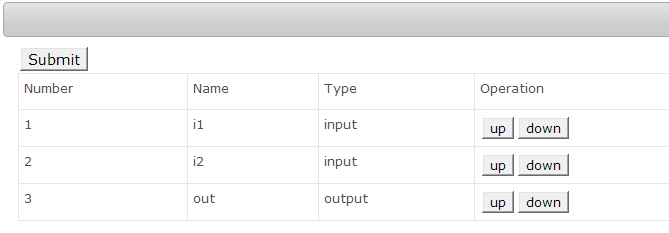
Param Order
When user click the “Param Order” button, the web page of adjust the order of command line parameters will be opened. Click the “Up” or “Down” the button to active it.
2)By XML format
BioCloud supports use XML format to load application.User can click “Xml Configuration Template” to view the template xml file.

2.3 Application share list

2.4 Application publish list
This page will show all the published applications of current user.When the application has been published, it can not be canceled.
